| Desired level of flexibility | Workflow requirements |
| Reference data inputs can be specified when submitting a job to GCE. | Parameters are unlocked in the workflow design. |
| Reference data inputs are pre-configured and cannot be overridden when submitting a job to GCE. | Parameter inputs in the workflow are locked. Separate workflows would be needed if the same analysis should be run using different reference data. |
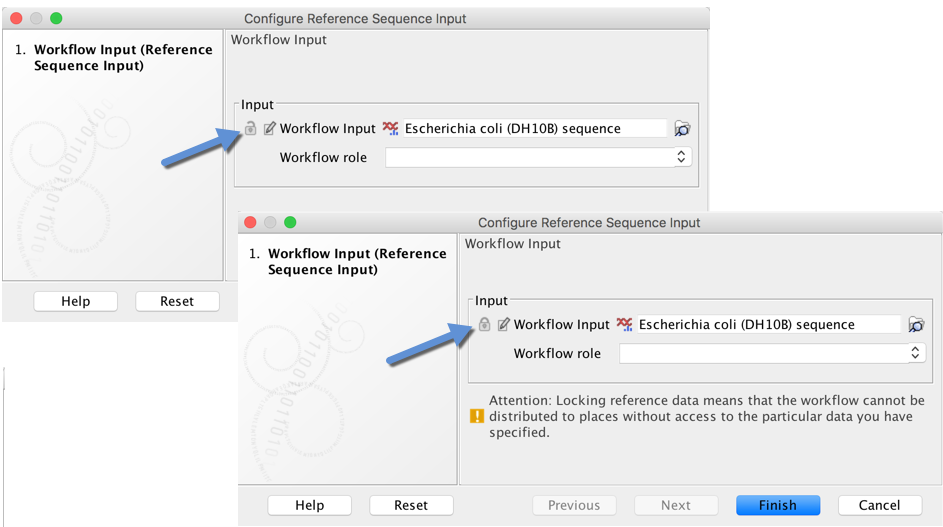 *Workflow parameter inputs can be configured as locked or unlocked. Values for unlocked parameters can be provided on the command line when submitting a job to GCE.*
## Creating a workflow installer
Information about creating workflow installers can be found in the Workbench manual at:
*Workflow parameter inputs can be configured as locked or unlocked. Values for unlocked parameters can be provided on the command line when submitting a job to GCE.*
## Creating a workflow installer
Information about creating workflow installers can be found in the Workbench manual at: 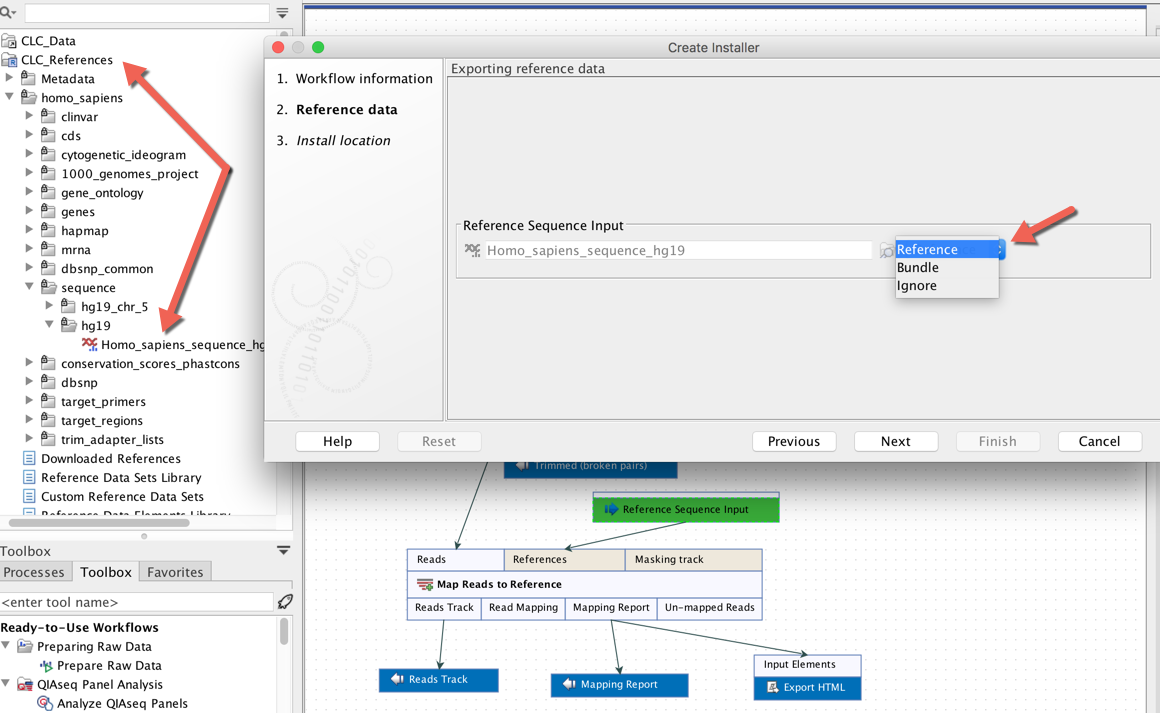 *When creating an installer for a workflow, up to three options will be provided for each data input as a parameter of a workflow element, "Ignore", "Bundle" and in some cases "Reference".*
*When creating an installer for a workflow, up to three options will be provided for each data input as a parameter of a workflow element, "Ignore", "Bundle" and in some cases "Reference".*
| Workflow installer option | What it means | Workflow design requirements | Recommended use |
| "Ignore" | Reference data is specified via the command line when submitting the job to GCE. The reference data must be uploaded to S3 prior to use. |
Workflow parameter inputs should not be configured with data elements and must be unlocked. | This is the standard way to work with reference data when running workflows on GCE. |
| "Bundle" | The reference data does not need to be specified on the command line. It is included within the workflow installer file. No separate upload of bundled reference data to S3 is needed. |
Workflow parameter inputs must be configured with a reference data element for the "Bundle" option to be available when making an installer. Workflow parameter inputs can be locked or unlocked. |
Can be useful when using very small reference data elements. |
| "Reference" | The reference data does not need to be specified on the command line. It is downloaded as needed when the workflow is run on GCE. It does not need to be uploaded to S3 manually. |
Workflow parameter inputs must be configured with a reference data element from a QIAGEN set for this option to be available when making an installer. Workflow parameter inputs can be locked or unlocked. |
Where this option is available, it is the most convenient option. |
 ````none
--export-bam-export-destination The destination to export the files to
````none
--export-bam-export-destination The destination to export the files to